We use systems biology, computational modeling, and biomolecular engineering approaches
to understand, predict and control genetic communication in bacterial communities
Systems & Synthetic Biology
Computational biology
Single cell quantification
Data Science & ML
Mathematical modeling
Biomolecular engineering
What's Comp Bio?
We love STEM, and we want others to feel the same. Not entirely sure what Computational Biology is? Check out our YouTube Channel!
We're Taking Questions
Have questions about Computational Biology that you'd like us to discuss? Check out our YouTube page or email us!
News
Check out what's new with the lab!
AICHE 2025
Congratulations to Chauner for placing 2nd in the General Engineering and Engineering Education category at the AIChE undergraduate poster presentation this year!

Ivan, Chauner, Varun, Arthur, and Will present posters at conferences this semester! 


AICHE 2025
Habib gives a talk at AIChE on his PlasAnn platform!
Congratulations to Habib for passing his qualifying exam! 
IMMEM XIV, 2025
Allison gives a talk at IMMEM this year!Congratulations to Zirui for passing her qualifying exam!! 
RMSC outreach event
The lab set up an interactive station at the Rochester Museum & Science Center (RMSC) during the Perseid Fest, as part of its annual outreach activity, welcoming visitors to explore and learn about Chemical Engineering and Microbiology!

Congratulations to Yuchang!
Congratulations to Yuchang for successfully defending her MS in Microbiology & Immunology!
2025 Microbial Population Biology GRS/GRC
Heather presents her work with a talk and a poster at the 2025 Microbial Population Biology GRS/GRC!Congratulations to Jordan!
Congratulations to Jordan on his graduation with an MS in Chemical Engineering! We wish him the best!!RES Young Engineer of the Year Award
Dr. Lopatkin was named a finalist for the Rochester Engineering Society's Young Engineer of the Year award for outstanding contributions to engineering practice in the Rochester region. Congratulations!
Publication in Nature Reviews Microbiology
A new review paper led by Mehrose exploring the role of bacterial metabolism in antimicrobial resistance!
New grant alert!
Off to a great start in 2025 with official award notice for the NSF CAREER grant to continue working on engineering HGT in microbial populations!
Happy holidays!!
Celebrating the end of the year holidays with our annual party at Allison's house featuring a spirited white elephant and lots of fun!!
New equipment alert!!
The lab purchases a bench-side FACS machine for better HGT quantification!
Congratulations to Abhishek!
Congratulations to Abhishek on his graduation with an MS in Data Science!

Publication in BioEssays
New review paper out led by Heather - beyond fitness costs, plasmids have many hidden advantages!
AICHE 2024
Varun, Habib, and Arthur represent the lab at AICHE 2024!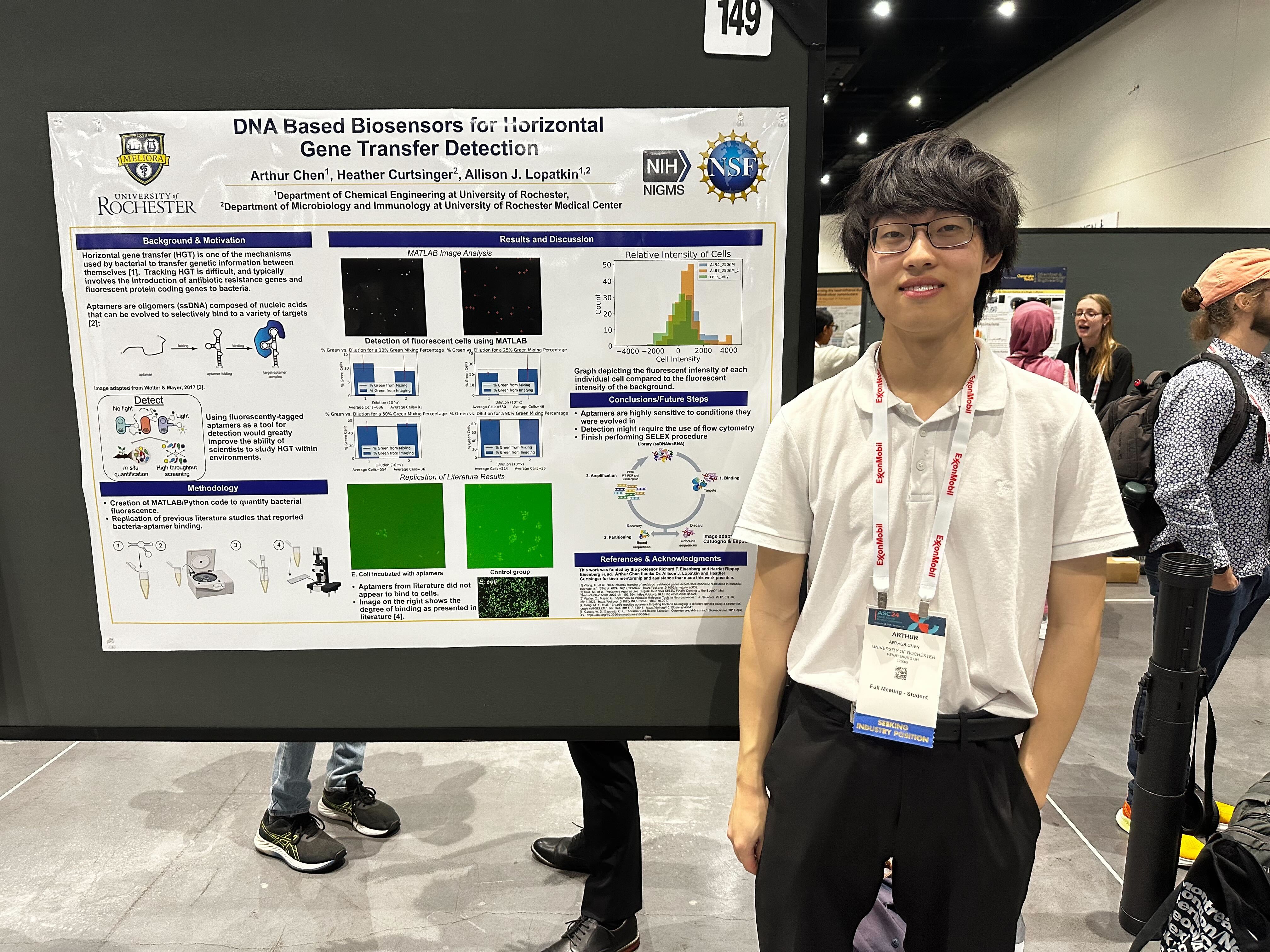


Publication in Microbiology Spectrum
New publication out showing that nucleotide synthesis is important in treating high density bacterial infections!
Eisenbergy symposium
Arthur presented his poster at the Eisenberg symposium as the lab's Eisenberg summer fellow!
Congrats to Heather and Varun for passing their qualifying exams!!
2024 Pew Scholar
Dr. Lopatkin is named a 2024 Pew Scholar (and is immensely grateful for it!!). More work on the intersection of metabolism and antibiotic resistance coming soon...
UR highlights Lopatkin's award
Excellence in Teaching
Congrats to PhD student Varun for winning the ChE Excellence in Teaching award!!
Pi Day 2024
The lab organized and led an interactive computational biology workshop with local Rochester city high school students in celebration of Pi Day! Students became microbiology forensic investigators and solved a Whodunnit?!: Bacterial bandit edition







Publication in Microbiology Spectrum
The lab's new work on antibiotic selection and metabolism is published in Microbiology Spectrum!
Research Talk
Varun gives a research talk at the University of Rochester AS&E Graduate Research Day 2023!
NIH R35
The lab receives NIH R35 funding to study determinants of horizontal gene transfer in pathogens
Edward Mallinckrodt Jr. Foundation Research Award
Dr. Lopatkin is thrilled to be a recipient of the Edward Mallinckrodt Jr. Foundation Research Award to study why certain pathogens pick up antibiotic resistance genes better than others!
Research News Feature
The lab's recent paper receives a mention!
Heather does the first experiment in the new lab!

Welcome to the lab!!
To all the new members: Sai Varun Aduru, Heather Curtsinger, Wenqi Di, Habibul Islam, Yuchang Liu, Elizabeth Martin, welcome!
We moved to Rochester!! And lab Construction is officially underway
The new lab in Gavett Hall is under construction! Here's a sneak peek...

New paper in Nature Communications
Rochester features our lab's recent paper!
Faculty News Feature
Check out our lab's recent feature on the Barnard faculty news page!
SRI Feature!
The lab was recently featured by Barnard in an SRI article- take a look to read about some of our members!
NIH Funding
The lab is excited to recieve NIH funding in collaboration with the Smith lab at NSU to study metabolism and antibiotic resistance!
Mehrose @ NY B.I.G.
Mehrose presents her work on drivers of plasmid acquisition costs at the NY B.I.G. conference!
New publication in J. Antimicrobial Chemo!
Mehrose's publication on carbapenem-resistant Enterobacterales is out in Journal of Antimicrobial Chemotherapy!
New publication in Microbial Genomics!
Claire's paper on Acinetobacter phylogenomics is out in print in Microbial Genomics!
Mehrose joins the lab!
The lab is excited to welcome our research technician Mehrose to the group! Read about her work here!
New round of funding!
The lab recieves its second federal funding from the NIH!
Publication in Current Opinion in Microbiology
Jenifer and Karolina publish their review on quantifying HGT in Current Opinion in Microbiology
